I finally tested my children’s DNA and got the results recently. This has been fascinating and I’m sure I’ll be writing more about it in the coming months. While working to interpret their tests, I created the common segment generator.
This new segment-based tool is helpful for a specific challenge that I encountered. I will concede that this is something of a niche situation, but I hope it will be helpful for some readers.
My use case
A couple of years ago, I completed a visual phasing project to figure out which segments I inherited from each of my four grandparents. I was able to cheat slightly and do a ‘turbo’ version of visual phasing. This is because as someone with parents tested, I could compare my phased maternal and paternal DNA with that of my three siblings. I completed the same process for my partner (who conveniently also has three siblings who were happy to test), resulting in the following maps.
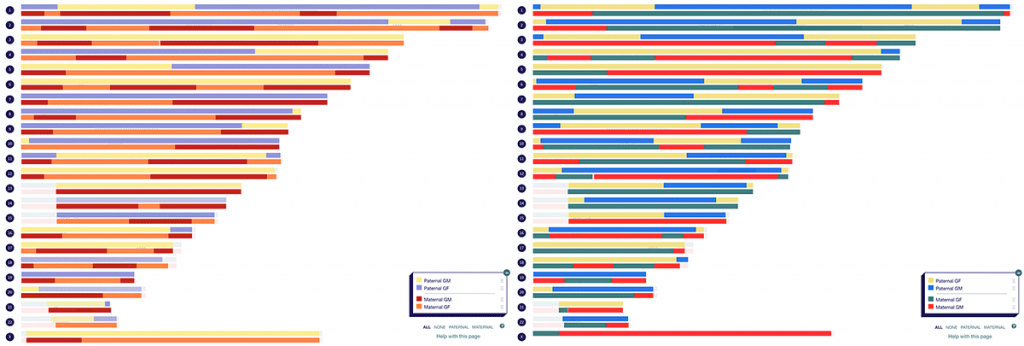
The first thing I did when the results came in for my children was compare their DNA with that of each grandparent. Exciting as this was, I knew that based on my phased map and that of their mother, I would be able to take my children’s maps to great-grandparent level. But how could I do this quickly and efficiently?
I needed to take the DNA each child shared with a grandparent and split it into DNA from each relevant great-grandparent. I had the data to do this, but no easy method. This is why I built this tool.
How it works
- There are text boxes for two sets of segments
- After you click ‘Generate Common Segments’, the tool will output a text box containing just the segments that exist in both sets
- It will recalculate the centimorgans for the common segments
- It uses a similar interface and method to the existing cM estimator tool
It’s not comparing any actual DNA; just the coordinates you give it. So it’s really only useful when you know the shared segments are on the same copies of a chromosome as in my use case.
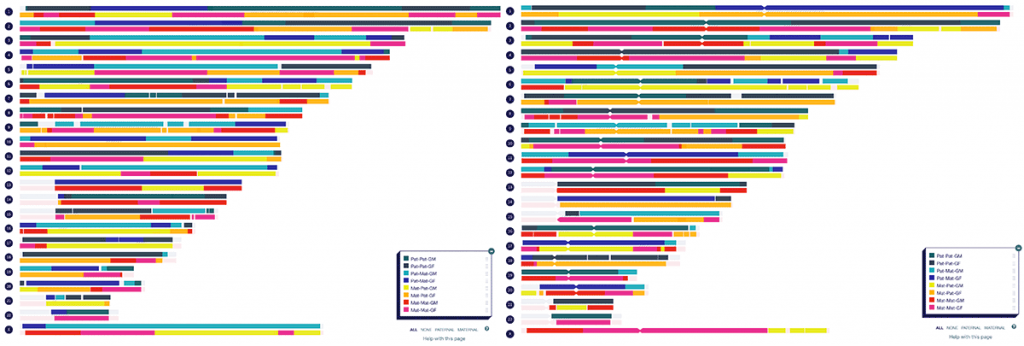
My children’s phased maps
To recap, I used the tool in two stages. First:
- I went to my phased chromosome map at DNA Painter, brought up the ‘All segment data’ report, and clicked ‘CSV’ to download a local copy
- I did the same for my partner, resulting in eight distinct groups of segments, each corresponding to one of my children’s great-grandparents
Then for each child, I went into the common segment generator. For each of their eight great-grandparents:
- In the first box, I pasted the segments that were assigned within the parent’s DNA for that grandparent (or great-grandparent to the child) during the phasing process described above. These represent all possible segments that the child could have inherited from that great-grandparent
- I then pasted the segments the child shares with the corresponding grandparent into the second box.
- After clicking ‘generate common segments’, the tool generated the segments that the child must have inherited from that great-grandparent.
- As a bonus, I now also have the approximate cM that they share with each of these great-grandparents!
The common segment generator tool
You can find the tool at http://dnapainter.com/tools/csg. I hope it’s helpful but realize not everyone will have a use for it.
You’ll notice there’s a segment threshold and that by default it’s set to a low 3cM. This is because for my use case I was using phased data with known relatives and did not want to exclude any valid segments. Even then, some obviously false segments came up on the X-chromosome that I had to delete.

If you find a novel use for the tool, please let me know!
Contact info: @dnapainter / jonny@dnapainter.com