As announced last month in the newsletter, the new distinct segment generator tool allows you to amalgamate a set of segments that more than one family member shares with a specific match. The tool will then output just the distinct segments, with the centimorgans (cMs) recalculated.
Why I built this
As a European tester without many significant matches, I find myself looking at dozens of connections around the 20-50cM mark. My challenge is that within this morass of matches:
- In many cases, the common ancestors will be from before the genealogical timeframe
- …while others are more recent and could potentially be traced
Identifying those that are traceable so that I can focus on building out their trees is an ongoing puzzle.
I have put a lot of effort into testing known relatives – e.g. siblings, cousins, the cousins of my parents. One possible method that occurred to me is comparing the segments that different known relatives share with a match:
- If they all share the same segment, then I might guess that perhaps this segment is from quite far back
- On the other hand, if across known relatives, there are multiple distinct matching segments, this makes it seem more likely that I’ll be able to find a connection.
For example, if my mother and her first cousin both share 40cM with a match across say two segments, it’s helpful if I can understand:
- If we’re looking at the same 40cM of shared DNA
- Or if in fact the distinct segments they share with this match amount to more than that altogether
- If they do, this could indicate that the connection is more recent
This approach isn’t a sure-fire way to identify traceable segments, but I thought it was worth a shot.
How the distinct segment generator works
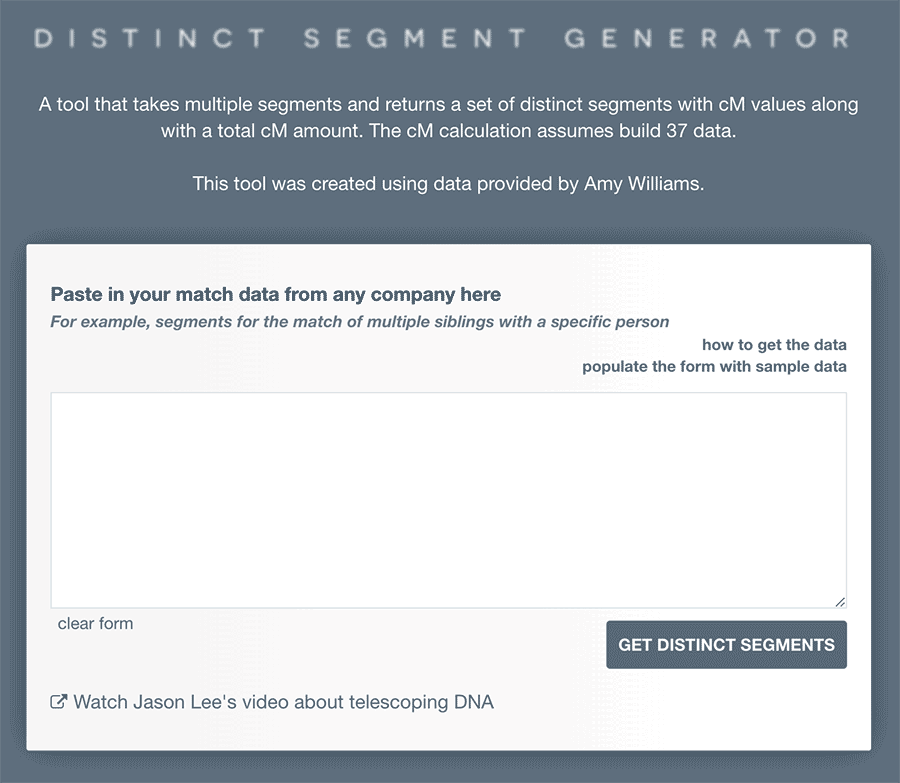
What you need
To use the tool, you’ll need the segment data for matches that both you and another family member share with a match. This means you’ll both need to match them on a site that has segment data (currently, all the main DNA testing sites except AncestryDNA and Living DNA). So this could for example be the segments that you share with a match, plus the segments a sibling shares with them.
Entering the data in to the distinct segment generator
You’ll see a familiar looking text box into which you can paste segment data. You can paste multiple sets of data into the same box. The tool will then return the distinct segments across all the matches.
Assessing the result
You can then consider the relationship based on different scenarios. For example:
- If the distinct segments amount to significantly more than the individual matches, then you can infer that your parent/grandparent shared more DNA with the match that you didn’t inherit, and the match may be worthy of more examination.
- Or if the distinct segments amount to no more than the individual matches, then the match might be less easy to trace (but of course if you have a lead on a match, it doesn’t mean you shouldn’t pursue it!)
The tool also has a link to a video that demonstrates another technique with which this tool could be used. I’ll also be releasing an variation on this tool in the coming weeks; please sign up to the newsletter if you’re interested.
This is an experimental tool I first put together last year (using the minimal genetic maps method in collaboration with Amy Williams). As always, I would be interested in all feedback, negative or positive!
Other blog posts on tools for cM calculations that may be of interest:
- Estimating cMs for a segment of DNA (cM estimator)
- More tips for inferred chromosome mapping (Inferred segments generator)
- Common segment generator
Contact info: @dnapainter.bsky.social / jonny@dnapainter.com
