Some answers to common questions are listed below. For feature-specific FAQs, please see the menu options.
I would be very grateful if you could email any suggested improvements or additions to this page to info@dnapainter.com.
General
DNA Painter is for anyone who would like visual tools to interpret results from an autosomal DNA test for family history research.
More applications for the site are emerging all the time. Here are a few ideas to start with:
- You can set up a map for each test result, visualising known ancestral segments as well as more speculative or unknown matches. You can make notes on segments and groups, and give an indication of how confident you are that you've identified a segment correctly.
- You could also make a quick map to illustrate a geneaological problem or puzzle, and then mark it as shared so that you can pass the URL around for review.
- You could take an ancestor (e.g. a grandparent or great-grandparent) and map all the segments their descendants share, mapping them in groups.
- Perhaps if you map what the segments for (say) a third cousin match might look like, it might pique the interest of the DNA match you're contacting!
I'm planning a page of case studies. Please get in touch if you have any suggestions or success stories to share.
In addition to the chromosome mapping tools, the site hosts general tools for genetic genealogy. The shared cM tool lets you enter the number of centimorgans you share with a match, and will then highlight possible relationships with probabilities for each. You can try this out here.
There's also 'What are the Odds'. There's a separate FAQ for that here.
And there's also a new feature allowing you to create or import an ancestral tree, which provides a nice-looking single-page summary of your family line. Read more
Common questions from new users
If you haven't already, it would make sense to upload your raw DNA to all sites who accept transfers. This currently includes Gedmatch, MyHeritage and FamilyTreeDNA (assuming you didn't test with either of the latter two). For more information on this, please visit 'What do you need to get started'.
You don't upload any raw DNA to DNA Painter. The site lets you 'paint' your own DNA by using the segments you share with your matches. When you create a blank chromosome map, this represents your chromosomes, with a maternal and paternal chromosome in each pair. You don't need to 'paint' yourself because the unpainted map already represents you.
You don't have to add yourself! A blank map contains a set of chromosome pairs that represent you. You can then start to map segments of your chromosomes to specific ancestors by 'painting' the segments you share with matches.
When you see segments overlapping on a chromosome, this doesn't necessarily mean that these people share the same segment of DNA. For each numbered chromosome pair, we all have both a maternal or paternal chromosome, but the comparisons made on Gedmatch and testing sites are not able to distinguish between the maternal and paternal chromosomes.
Imagine you have two matches, A and B, who seem to overlap on chromosome 4. You've compared yourself to both A and B, but until you've also compared A to B, you don't have a way of knowing that they actually share the same segment. It could be that A matches you on the maternal chromosome and B matches you on the paternal chromosome. The simplest way to triangulate a segment is by doing a one-to-one comparison on Gedmatch. If A matches B, then since you already know they both match you, the three of you share this particular segment of DNA. Depending on the size of the segment, this may mean you have a traceable common ancestor. If they don't match each other on this segment, then you know that you're looking at two distinct segments of DNA, one on your maternal chromosome 4 and one on your paternal chromosome 4.
If you are not able to do this on Gedmatch, you have a few other options when trying to determine if your two matches actually match each other on this segment:
- MyHeritage now have a chromosome browser that identifies triangulated segments
- FamilytreeDNA has a matrix tool. While this doesn't offer segment-level triangulation, it does give some indication (for example, if the ftDNA matrix tool does not show two matches as related, this is an indication that the overlapping matches are likely to be on different chromosomes)
For more information on triangulation, this post by Blaine Bettinger is highly recommended.
When you save a match to DNA Painter, this consists of one or more segments of DNA. The site will ask you for the name of the match. You should use a unique name (that is, if you have two matches called David, you should give them distinct names when painting their segments). Many users like to include the kit name of the match for future reference.
The site will also ask if you know how you're related to the match. If you do know how you're related, you should enter the name of the ancestor or ancestral couple from whom you deduce that you inherited this DNA (see assigning segments for more on this). If you don't know how you're related, you might want to group your matches who are related to each other with a relevant name, or perhaps use a name that narrows it down as much as possible (e.g. 'Unknown Paternal' or 'Unknown [Placename]).
On DNA Painter, colours are assigned to ancestors (or groups of matches), not to matches. The process of chromosome mapping aims to assign segments to specific ancestors - so it's these ancestors that are colour coded, not your DNA matches!
However, if you have an unknown match you can't place, you might prefer to have them in their own colour so that you can compare them more easily to the other segments you've painted. To do this, just add the match to their own group when you paint the segments (rather than choosing 'link these segments to an ancestor I've added before' and selecting an existing group).
To move an existing match to a new group, you have two options. First, click on any segment for the match you want to move, and click 'View match' on the popup. Then, either:
- Click 'Edit match'. A form will appear. You can check a box saying 'move all segments for ___ to a different group'. You can then create a new group.
- Or click and drag on the small dots to the left of the match's name on the overlay. After you start dragging, you will see a selection of boxes for all existing groups. You can simply drop the match into a different group, or onto the 'create new match' box.
DNA Painter *does not* know if your match is maternal or paternal! What the site can do is visualize the information you enter so you are better placed to figure this out yourself.
For example, when you paint a match, the site will point out the groups that contain segments that overlap with your match. In order to figure out if your new match actually triangulates with these segments, you will need to do a one-to-one comparison between the new match and the segments it overlaps.
For more info please see the FAQ item below, 'How do you find out if a match is maternal or paternal?'
This is set at the group level. So if you've painted a match in a paternal group and you want it to be maternal, you need to either:
- Move the match to a paternal group via the 'edit match' form
- Create a new paternal group via the 'edit match' form
- Edit the existing group to make it paternal
Chromosome Mapping aka DNA Painting
If you're lucky enough to have had one or more parents tested, you'll be able to tell based on which of your parents this person matches (or doesn't match) with. Beyond this, there are essentially two ways of figuring it out:
- If you know how you're related to the cousin (e.g. if you've done the research and found out who your common ancestors are - e.g. great great grandparents for a third cousin), then you'll know via which of your parents this relationship came.
- If you *don't* know how you're related to the cousin, you have to look for clues such as shared matches with other relatives you either know or have hunches about.
If a close cousin, aunt or uncle has tested, it might help to paint the segments you share with them as a guide. Then if a new match overlaps with your close relative, you can figure out if the new match is maternal or paternal by comparing them directly.
If you don't have the info you need to figure it out at this point, you have a few options that might help you to speculate. Some people have very distinct maternal and paternal heritage and are able to simply guess based on the location or ethnicity of their match (you can make notes to record your theories and make use of the certainty level if you're speculating). Or you can just select 'I don't know' from the dropdown asking you which side the match is on. Then in future when you have more evidence, you can move them onto the correct side.
If by any chance you're related to the cousin via both your mother and your father, it becomes a bit more complicated, but the process is essentially the same - you need to make use of other known matches in order to be able to place new ones - and that's essentially what the site is for!
Yes! One of the most important things to do in the early stages of interpreting your results is to find known relatives who have either tested, or would be prepared to test. Focus on the eldest generation first. You can also learn a lot from siblings, whose results can also be used for inferred matching and visual phasing.
The most useful matches for DNA painting are those you have with anyone who is more than one degree or generation removed. Closer matches can of course advance your research, but the segments you share won't allow you to distinguish between maternal and paternal segments, which is key.
These matches are very valuable to help determine how you're related to other more distant relatives. If you don't have a parent tested, it might be useful to paint the segments you share with these people in a pale colour as a guide.
- Close family, such as a full aunt or uncle, a half-sibling, or a parent: All segment matches you have with these people will get you over the first hurdle of interpreting your results, letting you know if the match is on your maternal or paternal side. There's probably not a lot of point in actually painting the segments you share with such a close relative; it makes more sense to use these people to determine how you're related to other more distant matches. However, if you don't have a parent tested, painting these segments could provide a useful guide; if a new match shares these segments, this will help you figure out if this match is maternal or paternal. Painting matches with a full sibling is not recommended (see "Should you add your mother, father or sibling to your map?" below); instead, start a separate map for your full sibling.
These matches will allow you to assign segments to specific ancestors, so you should paint them:
- A half uncle or aunt, or the first cousin of one of your parents, a second cousin, a great aunt or uncle, or a grandparent: All segment matches you have with these people can be attributed to a particular grandparent. For example, your second cousin is the descendant of a sibling of one of your grandparents, so you know that you inherited any common segments from that grandparent.
- More distant cousins: More distant matches can be very helpful - for example, you might share 100cM of DNA with a third cousin, all of which you can paint as coming from one particular great grandparent.
- Groups of unknown cousins: You may find within your DNA matches that a particular ancestor name keeps coming up that is unknown to you. Whether or not you are able to figure out any connection to this person, it can be helpful to paint a group of these segments for future reference.
All known matches are useful - even remote cousins - but the closest ones are considerably more useful. For example, you might share around 500 cM with a first cousin once removed and 850cM with a half uncle or first cousin. Once you have the data for these matches, any future matches can be compared against them, so unless you are related to them in more than one way (or are contending with a lot of endogamy):
- if someone matches you and your paternal first cousin, they are a paternal match
- if someone matches you and your mother's paternal first cousin (your first cousin once-removed), the matching segments came from your maternal grandfather
- if someone matches you and your half-uncle (your father's half-brother via his father), the matching segments came from your paternal grandfather.
You might need to speculate along the way. DNA Painter includes a 'certainty level' and notes fields to help you do this.
In this case it's more difficult! But the situation could change very quickly as more and more people test.
When you first start out, the number of unknown matches might seem overwhelming, so it's worth remembering:
- Once you start to identify matches, or even just narrow them down to a parent or grandparent, you'll have much more to go on when new matches come in
- Just focus on the significant matches first. What's significant will depend on your results, but a common marker is a match where there's a segment of at least 20cM or thereabouts.
It probably doesn't make sense to start chromosome mapping until you have at least one match identified.
While there's nothing to stop you doing this, if your aim is to map segments to ancestors, there's no real value in adding the segments you share with a parent or sibling to your map, since:
- The DNA we get from our fathers fills all the paternal chromosomes, and the DNA we get from our mothers fills all the maternal ones
- Any segments you share with a full sibling will be a mixture from your maternal and paternal chromosomes, and you won't have any way to distinguish these (unless you have phased both kits - read more here).
- Even if you have a half-sibling and can therefore paint the matches onto either the maternal or paternal chromosomes, doing so doesn't help you to map any segments to ancestors since the common ancestor in this case is one of your parents!
For close relatives like this it makes sense to set up a separate map for each. Also be sure to investigate inferred matching.
This site lets you assign segments to ancestors by making use of the segments you share with known matches. To assign segments to ancestors, you need to know how you're related to someone with whom you have a DNA match.
For example, say you match a second cousin called Bill. Your most recent common ancestors are your great grandparents Robert and Susan. The process is then quite simple: you can infer that you inherited the segments you share with Bill from these common ancestors.
As a cautionary note, it's worth mentioning that if your ancestry includes an endogomous community, or if your parents are quite recently related to each other, this becomes more complicated.
As noted above, you can generally infer that the segments you share come from the couple who are the most recent ancestors you share with your match.
Some people undertaking chromosome mapping like to use the name of this ancestral couple as their group name - e.g. Robert and Susan Smith. To assign them to an individual (which is my personal preference), you'd simply go forward one generation and name your group after the grandparent you're descended from (in my example this would be the child of Robert and Susan Smith). There's an additional discussion of this here with a diagram.
It's a personal choice, but most people would take an approach along the lines of the following. Say you have a DNA match where they are your third-cousin twice:
- If you consider your aim is to assign segments of your DNA to your ancestors as specifically as possible: with a normal third cousin relationship you can infer 'this DNA came from this specific set of great-great grandparents.' But in this case you'd have to say 'these segments could have come from any of these two sets of great-great grandparents'
- So the most common approach would be to enter all four last names for now (e.g. each of the possible sources of DNA). In future you may find evidence (e.g. cousins you're only related to on a specific branch) who can help you assign the segments more specifically. In these cases you can edit the segment individually and move it to a more specific group.
Click on any segment for the match, and click the 'view match' button. You'll then see a link at the top of the overlay: 'Add new segments for [match name]'. This can be helpful when adding smaller segments or X-segments that weren't in your initial set of segments.
When you create a new group, the site will suggest a random colour, but you're free to choose whichever colours you like.
It obviously makes sense to have a method, such as choosing four distinct colours for each of your grandparents, and then using similar colours for other ancestors within each quadrant. You're also likely to identify lots of matches where you can narrow them down to a certain degree (e.g. to a grandparent or parent), but no further. Again, using a colour for an 'unknown [grandparent]' group that's related to the colour you used for the grandparent themself can help when analysing segments.
Inferred matching is a process by which you can use the relationship that a close relative has with a known cousin to infer the source of the equivalent segments on your own chromosomes. For example:
- Parent: The DNA you share with your mother and father will fill the whole of the maternal or paternal side of each chromosome pair. However, if you have access to the tests results of one of their cousins, you can use these to narrow down lots of additional segments; for example:
- The segments that you share with your mother's paternal cousin (the child of one of your mother's father's siblings) can be attributed to your maternal grandfather
- The segments that your mother shares with this cousin, but which you do not share with this cousin, are inferred matches: because you inherited all your maternal-side chromosomes from your mother, but you don't share these segments with the paternal cousin, you know that you got these segments from your mother's mother (if you'd got them from her father, you would also match on them with the paternal cousin).
- Sibling: Similarly, if you have any known matches, the difference between the segments your siblings share with them and the segments you share with them can be used for inferred matching.
- A word of caution: inferred matching is inevitably slightly speculative, since there may be genuine segments that you share with the match, but which are so small that they don't fall within the typical 7cM threshold. Thus it's possible to assign some segments incorrectly via this technique.
For a more detailed set of examples, please read this article on using the site with inferred matches.
Using the site
You need the 'Match segment data' - a list of the chromosomes and segments you match on with a particular person.
For instructions and screenshots for each vendor, please visit the page Where do I find the match data?
To edit the name, gender or description, simply click on the text. It will transform into an editable field and will save when you click away from it.
- Segments You can edit an individual segment by clicking on it and clicking 'Edit Segment'. This then lets you edit the match name, confidence level, start, end, cMs and SNPs, as well as switching the group and adding segment-based notes. Within this edit form you'll also see buttons allowing you to delete a segment or make a copy of it (this can be useful if you want to split it across more than one ancestor). You can also delete several segments at one time as follows:
- Click on the settings cog above chromosome 1
- Click 'all segment data'
- Click 'mark for deletion'
- Check the box to the left of the segments you want to delete, and click 'Delete checked segments'
- Matches You can edit a match by clicking on a segment and clicking 'Edit match' (you can also access this form from the match overlay). You can move all match segments for a match to a different group, set their confidence level, and write notes to be applied to all of them. You can also delete a match and all its segments by clicking the 'Delete Match' button at the bottom left of the edit match form and confirming. Please note: matches are identified using the match name - so if you have duplicate segments for a match, you'll need to delete these individually. You could alternatively use the 'Remove duplicate segments' tool within the settings cog, or delete the entire match and reimport.
- Groups To edit a group, click on its name in the key to bring up the group overlay, and click the 'Edit Group' button on the right. You can also reach this overlay by clicking on any segment and clicking 'View entire group'. Within the group you can edit the name, colour and group notes, as well as setting a confidence level for all segments in the group. You can also delete the entire group if you need to by clicking 'Delete Group' at the bottom left of the 'Edit Group' form. You can also merge a group into another one by clicking the button at the bottom left of the group overlay and selecting a group to merge into.
- Group and layer order You can use the dotted 'grabber' area to the right of each group in the key to drag and drop the groups and change their order in the key. This order will then also alter the layer order of segments throughout your map.
To delete a map, go to your dashboard click the 'X' at the top right of the map you want to delete. You will be asked to confirm before deletion.
As you assign more segments, you might find that you want to split existing segments in order to assign parts of them to further back ancestors. To do this you have two options:
- Don't bother (!) - just re-order the groups with the furthest back ancestor group on top and let the site layer the segments for you.
- Manually split the segments. To split a segment, click 'Edit segment'. You'll see a button next to the save button called 'Copy Segment'. When you click this, a popup will appear, confirming what will happen next. You can then make a copy and manually adjust the start/end points as required.

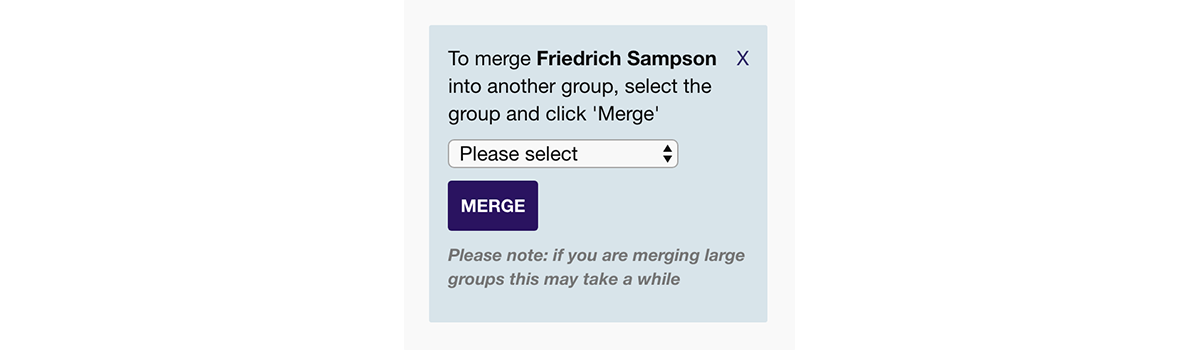
You may occasionally find you want to merge two sets of grouped segments. To do this:
- In the key at the bottom right of your map, click on the group that you want to merge into another group. This will bring up the group overlay.
- Within the group overlay, you'll see a button a the bottom left, 'Merge into another group'. If you click this, a select box with a list of your other groups will appear.
- Select the group you want to merge into, and click 'Merge'. You will then be taken to the new merged group.
To do this, export the segments from one map and import them into another using the 'Import segment data' tool:
- In the settings area of the first map, click 'All segment data' under 'Reports'
- If necessary, filter the table for the segments you want (skip this step if you want all segments)
- Click 'CSV file' at the top left, and a CSV file will be downloaded
- In the other map, go to 'Import segment data' within the settings area above chromosome 1
- Select your previously downloaded CSV file, adjust the thresholds as necessary. and click 'Import this file'
If you click on the settings button (the 'gear' cog just above chromosome 1 on the map page), you'll see an option to mark the map as shared. If you click the switch to make the map shared, the site will display a special share link. Anyone who has access to this link will be able to view (but not edit) your map.
This is the certainty level in action. The less certain you indicate you are in a match, the paler it will be displayed.
These are 'pile up regions'. The site includes them as a guide to some of the areas that have so far been identified by ISOGG as being prone to what they call "excessive IBD sharing". Effectively this means that more people share DNA at these points than would normally be expected, so if you share DNA with matches only on these sections, the match might not be recent enough to be traceable. Read more on the ISOGG website.
If there's an image associated with your email address, it will appear next to your name at the top right of the navigation bar. To associate one, go to https://gravatar.com/ and upload an image or icon.
Shared cM Tool with Probabilities
For more information, please read this explanatory blog post.
Privacy and Security
- The data you enter is the 'match segment data' as opposed to your actual genetic results or raw DNA. So for example if you and I matched, you'd be pasting in the information about where on the chromosome we match (the chromosome number, the start point, the end point, the number of centimorgans and SNPs), as opposed to the values that we match on (e.g. AATG etc)
- A lot of work has gone into making the site as safe and secure as possible. Communication between you and the site is encrypted and authenticated using TLS 1.2, ECDHE_RSA with P-256, and AES_128_GCM. That said, it's notoriously difficult to make any site 100% secure, you should store only the information you need for your research on the site.
- DNA Painter is a web-based application as opposed to a program you download, and the data you enter is saved in the site's online database. It can only be accessed by you, unless you choose to make your map 'shared', in which case it can be accessed by anyone you give the link to.